IN A NUTSHELL
1. About This Work
Type III secreted effectors (T3SEs) can be injected into host cell cytoplasm via type III secretion systems (T3SSs) to modulate interactions between Gram-negative bacterial pathogens and their hosts. Due to their relevance in pathogen-host interactions, significant computational efforts have been put toward identification of T3SEs and these in turn have stimulated new T3SE discoveries. However, as T3SEs with new characteristics are discovered, these existing computational tools reveal important limitations: (1) most of these trained machine learning models, based on the N-terminus (or incorporating also the C-terminus) instead of the complete sequences, and (2) the underlying models (trained with classic algorithms) employed only few features, most of which were extracted based on sequence-information alone. Thus, to achieve better T3SE prediction, we must identify more powerful, informative features and investigate how to effectively integrate these into a comprehensive model.
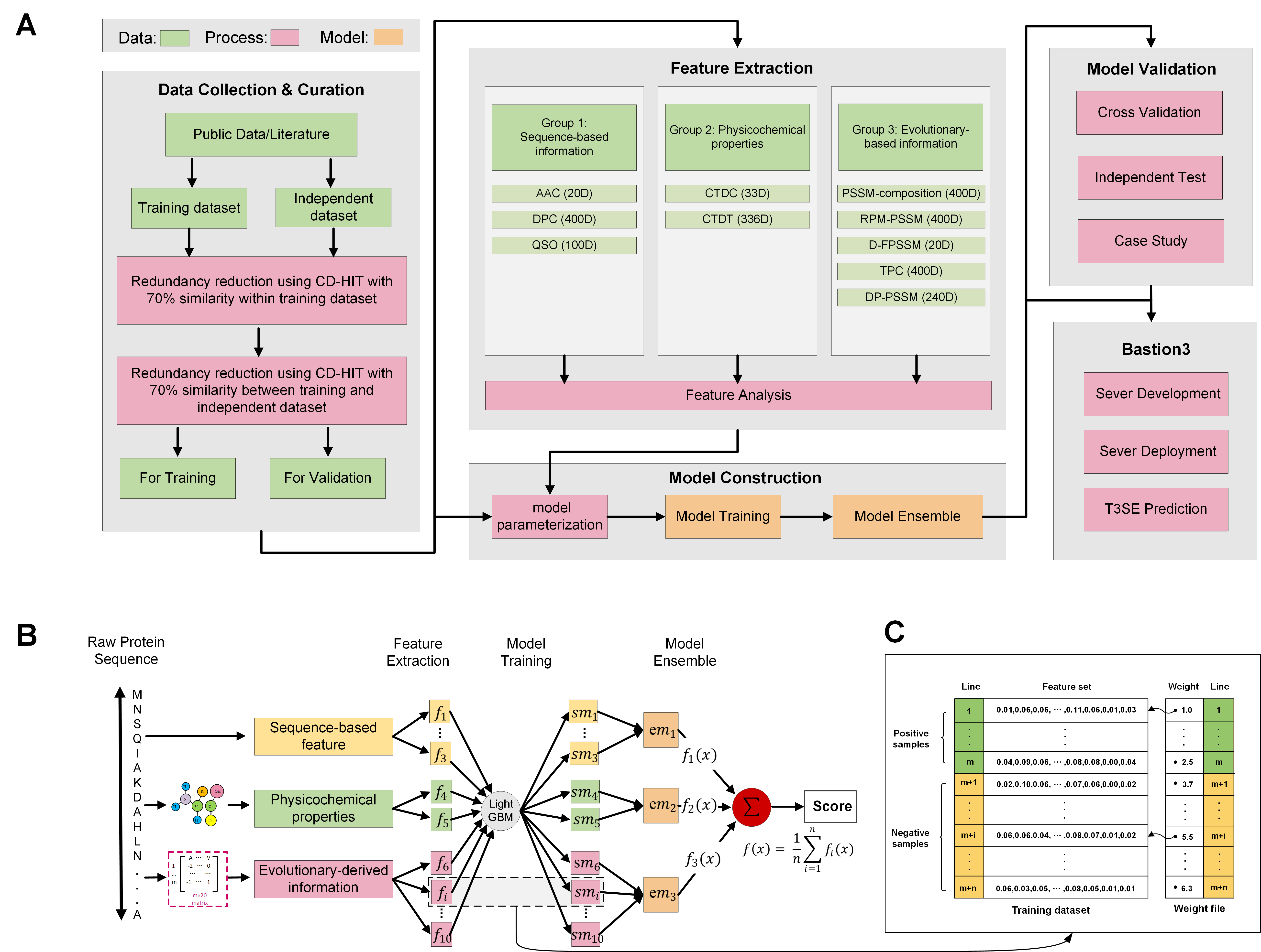
In this work, we present Bastion3, a two-layer ensemble predictor developed to accurately identify type III secreted effectors from protein sequence data. In contrast with existing methods that employ single models with few features, Bastion3 explores a wide range of features, from various types, trains single models based on these features, and finally integrates these models through ensemble learning. Specifically, we trained the models using a new gradient boosting machine, LightGBM, and further boosted the models' performances through a novel genetic algorithm (GA) based two-step parameter optimization strategy. Our benchmark test demonstrates that Bastion3 achieves a much better performance compared to commonly used methods, with an ACC value of 0.959, F-value of 0.958, MCC value of 0.917 and AUC value of 0.956, which comprehensively out-performed all other toolkits by more than 5.6% in ACC value, 5.7% in F-value, 12.4% in MCC value and 5.8% in AUC value. Based on our proposed two-layer ensemble model, we further developed a user-friendly online toolkit, maximizing convenience for experimental scientists toward T3SE prediction. With its design to ease future discoveries of novel T3SEs and improved performance, Bastion3 is poised to become a widely used, state-of-the-art toolkit for T3SE prediction.
DATASETS
1. Construction of the Training Dataset
We constructed the training dataset by mining currently known effectors from the literature, as well as cross-referencing to several existing T3SE datasets (An, et al., 2017; Arnold, et al., 2009; Dong, et al., 2015; Dong, et al., 2013; Samudrala, et al., 2009; Tay, et al., 2010; Wang, et al., 2013; Wang, et al., 2011; Yang, et al., 2013), and non-effectors from previous works (Wang, et al., 2017; Wang, et al., 2018). After manually removing wrongly annotated effectors and homologous sequences at the threshold of 70%, clustered by the CD-HIT program (Huang, et al., 2010), the final dataset contained 379 effectors and 1112 non-effectors.
2. Construction of the Independent Test Dataset
We subsequently generated an independent test dataset by manually extracting T3SEs from recently published literature and non-T3SEs from various bacterial species, in order to rigorously evaluate the predictive capability of our proposed method, and compare it against the existing state-of-the-art T3SE predictors. After removing highly homologous samples (with more than 70% similarity) from our training dataset, we finally constructed the independent test dataset containing 108 T3SEs and 108 non-T3SEs.
3. Case Study sequences
We further performed a case study, using additional three very recently exper-imentally validated T3SEs and examined in detail the predictive performance of different approaches.
ONLINE WEB SERVER
1. Bastion3
To maximize the users' convenience without going through the complicated algorithmic details, we have developed a user-friendly and easy-to-use web server, termed Bastion3, as an implementation of the proposed two-layer ensemble approach.
Please note the following important aspects of the Bastion3 web server:
2. Using the Bastion3 web server
Bastion3 is an online server implemented with a user-friendly interface, which makes it very easy to use. All you need to do is to fill the input sequence box or upload a sequence file. Upon its submission, the prediction job will be placed in the queue system. All the submitted jobs will be processed by the Bastion3 server successively. After your job is finished, you will receive an e-mail with a URL of your job results if you provide an email address.
2.1 Input Formats
Two types of input are accepted by Bastion3: sequences in FASTA format (strongly recommended) and raw sequences.
In the case of input sequences in the FASTA format, you can prepare and input them as follows:
In addition, the following input sequence, which is in the original format downloadable from the UniProt database:
will be formatted (in order to remove those line breaks within the sequence) as follows:
In the case of raw sequences, you can input them as follows:
which will be formated by Bastion3 as follows:
2.2 Input sequence limits
1. The length of each submitted sequence should be in the range of 31 and 5000.
2. Considering that T3SE prediction is a little bit time-consuming, the maximum number of sequences allowed for each submission by the Bastion3 server should be no more than 500.
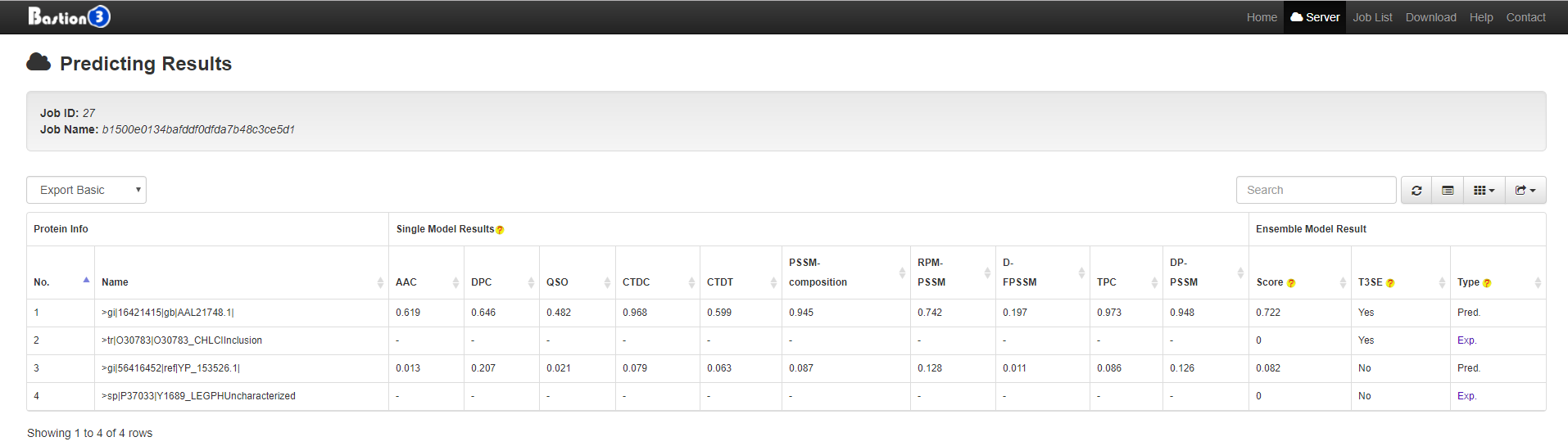
3. Bastion3 Prediction Result Instructions
There exists a built-in list (continuously updated to keep in pace with BastionDB) of major types of secreted effectors (such as types II, III, IV and VI secreted effectors) in Bastion3 to annotate the prediction results after jobs are processed, through which we aim to distinguish the known effectors from the computationally predicted ones and provide detailed annotations of those known effectors for users.
For a computationally predicted secreted effector, the results are marked as Pred, while the detailed prediction results (including those predicted by the single method-based models and those predicted by the final ensemble model) will also be presented to users.
For a known secreted effector (such as the protein O30783), the results are marked as Exp, and a corresponding URL link to the BastionDB will be provided to users that contain detailed information on this effector (an example is provided in the following figure).